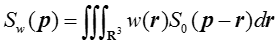CLARITY-based tractography for projection tracking
Tractography methodology akin to what is used for diffusion MRI was used to reconstruct 3D models of fiber bundle trajectories based on the CLARITY data. The tractography algorithm 52 propagates streamlines from a “seed” region through a vector field of voxel-wise principal fiber orientations and terminates if a streamline makes a sharp turn (angles larger than a prescribed threshold αthresh) or ventures outside of the masked region. For each voxel, the principal fiber orientation was estimated from a structure tensor, which was computed using the image intensity gradients (as a marker of the edges of fiber tracts) within a local neighborhood of the voxel53-58. The principal fiber orientation was defined as the tertiary eigenvector (i.e. with the smallest eigenvalue) of the structure tensor. The structure tensor was defined as:
where p and r represent spatial locations, w is a Gaussian weighing function with standard deviation σg, S0 is a symmetric second-moment matrix derived from image intensity gradients:
where Ix, Iy and Iz are the gradients of image intensity I along each of the x, y and z axes, computed by convolving I with three 3-dimensional 1st order derivative of Gaussian functions of standard deviation σdog59. Structure tensors were computed using MATLAB software (MathWorks, Inc.)
Tractography and tractography visualization were performed using Diffusion Toolkit and TrackVis softwares (http://trackvis.org/) respectively.
Matlab code can be download here
Representative intermediate steps of reconstructing projection to streamlines using structural tensor based CLARITY tractography.
a, Raw CLARITY image, showing outgoing mPFC projections (EYFP). b, Image intensity gradient amplitude, computed by convolving the 3-dimensional CLARITY image volume with three 3-dimensional 1st order derivative of Gaussian functions, c, Color-coded principal fiber orientations (A-P: red; D-V, green; L-M, blue), estimated as the tertiary eigenvectors of computed structure tensors. For better visualization, the color brightness was weighted by the raw CLARITY image intensity. Scale bars in a-c: 100 µm. d, A zoomed-in region of (c) showing the principal fiber orientations as color-coded vector fields overlaid on raw CLARITY image. The vectors are color-coded for their orientation. Scale bar: 6µm.
Step-by-Step Tractography guide:
--coming soon